|
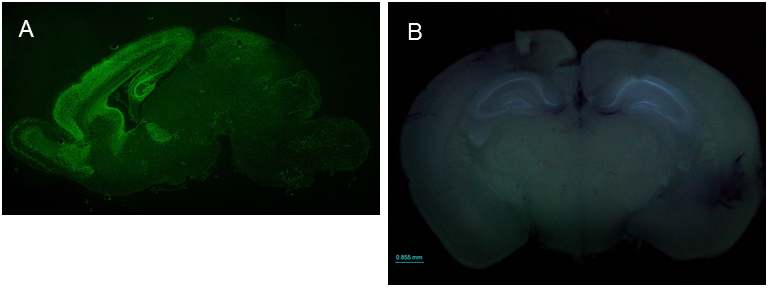
SIP1 expression patterns in the brain ICR;129(Cg)-Zfhx1b<tm2.1Yhi> (RBRC09239)Courtesy of Yujiro Higashi, Ph.D. (A) Immunofluorescence staining of a sagittal section of the SIP1-EGFP mouse brain at P0 stage using anti-GFP antibody shows that SIP1 (SIP1-EGFP) is expressed in the cortex, hippocampus, olfactory bulb, ventricular zone of the lateral ventricle and cerebellum. (B) Fluorescence emission observed in the freshly prepared brain from 8-week old SIP1-EGFP mouse shows that SIP1 is clearly expressed in the hippocampus. SIP1 expression in the hippocampus seems to continue throughout life. |
| Mowat-Wilson syndrome (MWS) is caused by de novo heterozygous mutation or deletion in Smad-interacting-protein 1 (Sip1, also known as Zeb2/Zfhx1b) at 2q22. MWS is characterized by distinctive facial appearance, intellectual disability, epilepsy, and other features including Hirschsprung disease and heart defects [1]. Sip1 homozygous null mutant mice show an embryonic lethal phenotype with a variety of defects at embryonic day 8.5. SIP1 is a transcription factor and also interacts with Smad proteins, regulating multiple signaling pathways. Previous studies reported that SIP1 plays an important role in neuron and glial cell differentiation, central nervous system myelination, and migration of the cortical interneurons [2-4]. Higashi and colleagues generated a SIP1-EGFP knock-in mouse strain to detect SIP1 protein in vivo and revealed that EGFP expression was observed in oligodendrocytes of the corpus callosum and fimbria, olfactory bulb, Bergmann glial cells of the cerebellum, and raphe nucleus of the brainstem, as well as in the hippocampus and cerebral cortex [5]. SIP1-EGFP mice provide the opportunity to clarify the physiological and pathological roles of SIP1. |
| Depositor | : | Yujiro Higashi, Ph.D. Department of Perinatology Institute for Developmental Research, Aichi Human Service Center |
|
| Strain name | : | ICR;129(Cg)-Zfhx1b<tm2.1Yhi> | |
| RBRC No. | : | RBRC09239 | |
| References | : | [1] | Mowat DR, Wilson MJ, Goossens M. Mowat-Wilson syndrome. J Med Genet.; 40(5):305-10, 2003. |
| [2] | Seuntjens E, Nityanandam A, Miquelajauregui A, Debruyn J, Stryjewska A, Goebbels S, Nave KA, Huylebroeck D, Tarabykin V. Sip1 regulates sequential fate decisions by feedback signaling from postmitotic neurons to progenitors. Nat Neurosci.; 12(11):1373-80, 2009. | ||
| [3] | Weng Q, Chen Y, Wang H, Xu X, Yang B, He Q, Shou W, Chen Y, Higashi Y, van den Berghe V, Seuntjens E, Kernie SG, Bukshpun P, Sherr EH, Huylebroeck D, Lu QR. Dual-mode modulation of Smad signaling by Smad-interacting protein Sip1 is required for myelination in the central nervous system. Neuron; 73(4):713-28, 2012. | ||
| [4] | McKinsey GL, Lindtner S, Trzcinski B, Visel A, Pennacchio LA, Huylebroeck D, Higashi Y, Rubenstein JL. Dlx1&2-dependent expression of Zfhx1b (Sip1, Zeb2) regulates the fate switch between cortical and striatal interneurons. Neuron; 77(1):83-98, 2013. | ||
| [5] | Nishizaki Y, Takagi T, Matsui F, Higashi Y. SIP1 expression patterns in brain investigated by generating a SIP1-EGFP reporter knock-in mouse. Genesis; 52(1):56-67, 2014. | ||
| July 2015 Contact: Shinya Ayabe, Ph.D. Experimental Animal Division, RIKEN BioResource Center All materials contained on this site may not be reproduced, distributed, displayed, published or broadcast without the prior permission of the owner of that content. |