1.Overview:
The lymphocytic choriomeningitis virus (LCMV) causes either acute or persistent infection in rodents, and opportunistic infection in humans. In order to detect LCMV infection in our mouse colony, we have established a protocol for RT-PCR test which can amplify the nucleoprotein (NP) region of LCMV small (S) RNA using RNA extracted from mouse kidneys.
2.Background:
RIKEN BRC and the National Institute of Infectious Diseases isolated the LCMV which infected the MAI/Pas mouse colony, then determined whole sequences of S RNA. Using this sequence information and the reported LCMV S RNA sequences found on the public databases, we designed the degenerate primer set written below which can amplify the NP region of the isolated LCMV, LCMV Armstrong and LCMV WE strains and set the reaction condition.
| Primer: | NP5-001 | 5′-tccatragwgcacagtgyggggtgat-3′ |
| NP3-001 | 5′-gcatgggaraayacracaattgayc-3′ | |
| r =a or g, w = a or t, y =c or t | ||
3.Materials and methods:
1) RNA extraction
RNA was extracted from 5 mg of kidney tissues using a QuickGene RNA tissue kit S (Fujifilm Co., Ltd., Tokyo, Japan) by a beads cell disrupter (ZB-50 and MicroSmash MS-100R, Tomy Seiko Co., Ltd., Tokyo, Japan) and automated extraction system (QuickGene-800, Fujifilm Co., Ltd,. Tokyo, Japan). Concentration of the finally obtained RNA was 500 to 1,000 ng/μl.
2) cDNA synthesis
cDNA was synthesized using SuperScript II (18064-071, Invitrogen Corp., Carlsbad, CA, USA) and random primer (C1181, Promega Corp., Madison, WI, USA).
Reaction mixture:
| ddH2O | 1.25 μl | ||
| 5 x Buffer | 2.0 μl | ||
| dNTP (2.5 mM) | 1.25 μl | ||
| DTT (0.1 M) | 1.0 μl | ||
| Random primer (500 μg/ml) | 0.5 μl | ||
| Extracted RNA | 3.0 μl |
Reaction mixture above was prepared and incubated at 70°C for 3 minutes, at 55°C for 90 seconds and at 37°C for 1 minute. One μl of SuperScript II was then added to the mixture to make the final volume of 10 μl. cDNA was synthesized by the incubation of the mixture at 37°C for 20 minutes and at 98°C for 5 minutes.
3) Preparation of PCR reaction mixture
QIAGEN Multiplex PCR kit (131965, QIAGEN Inc., Hilden, Germany) was used. The PCR reaction mixture was as follows:
| ddH2O | 6.0 μl | ||
| primer: NP5-001 (10 μM) | 1.0 μl | ||
| primer: NP3-001 (10 μM) | 1.0 μl | ||
| synthesized cDNA | 2.0 μl | ||
| 2 x QIANGE Multiplex Master Mix | 10.0 μl |
4) PCR reaction
The detection of LCMV was performed using a degenerate primer set of NP5-001 and NP3-001 described above which amplified the NP region of LCMV S RNA. Control PCR was executed to ascertain the RNA extraction and cDNA synthesis using a primer pair that amplified glyceraldehyde-3-phosphate dehydrogenase (G3PDH, PCR-301, Toyobo Co., Ltd., Osaka, Japan).
PCR condition:
| 95°C | 15 minutes | ||
| 94°C | 30 minutes | ||
| 60°C | 90 minutes | x 30 cycles | |
| 72°C | 90 minutes | ||
| 72°C | 10 minutes |
5) Electrophoresis
Ten μl of PCR products were applied to agarose gel electophoresis. Electrophoresis was performed on 2% agarose gel in 1 x TAE buffer for 100V, 30 minutes, using one hundred base pair ladder marker (15628-050, Invitrogen Corp., Carlsbad, CA, USA).
4.Results:
The typical results are shown below (Figure 1). RT-PCR clearly demonstrated that only MAI/Pas mice carried LCMV.
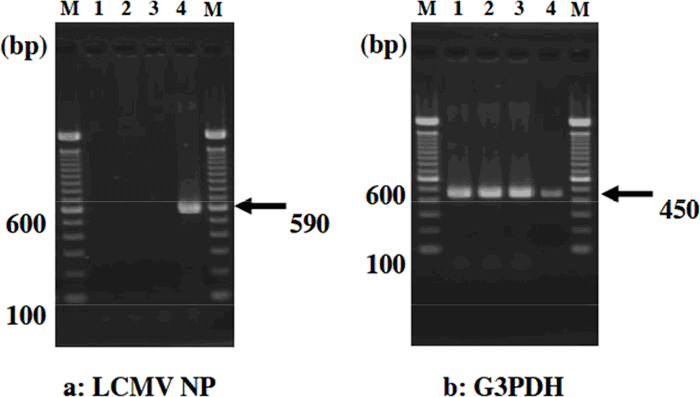
Figure 1. M: one hundred base pair ladder marker (15628-050, Invitrogen Corp., Carlsbad, CA, USA)
Lane 1:Strain A (LCMV negative), Lane 2: Strain B (LCMV negative)
Lane 3:Strain C (LCMV negative), Lane 4: MAI/Pas (LCMV positive)
5.Conclusion:
We could establish a reliable RT-PCR test to detect LCMV NP region in a persistently-infected mouse.
Acknowledgement:
We acknowledge Dr. Shigeru Morikawa at the National Institute of Infectious Diseases, Tokyo, Japan for his overall cooperation in the virus isolation, S RNA sequence and RT-PCR.
Fumio Ike, Ph.D.,
Hatsumi Nakata, Ph.D.,
Atsushi Yoshiki, Ph.D.
Experimental Animal Division
RIKEN BioResource Center




