|
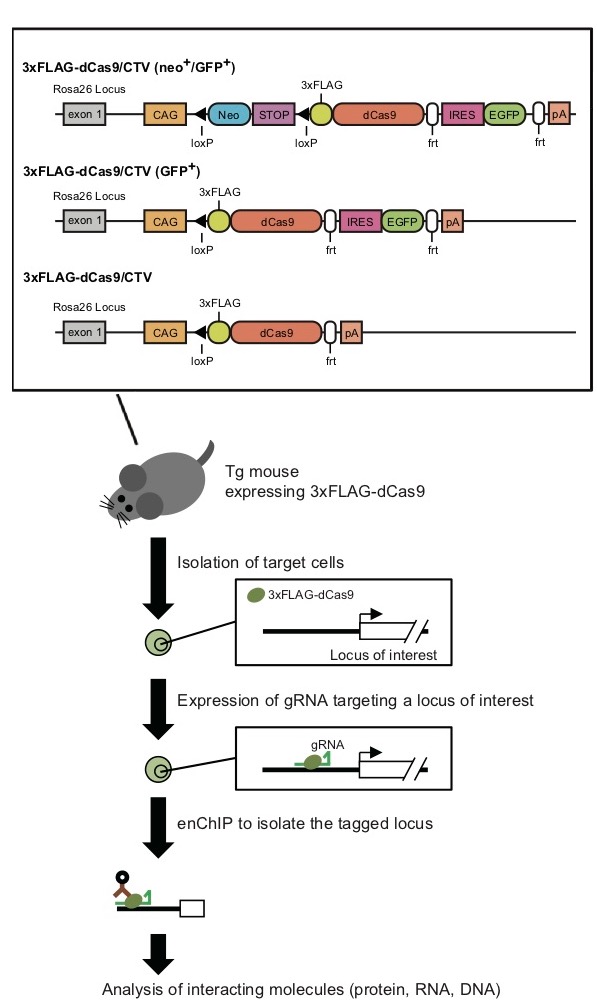
3xFLAG-dCas9 expressing mice for enChIP analysis C57BL/6-Gt(ROSA)26Sor<tm1(CAG-3XFLAG/dCas9,-EGFP)Hfuj> (RBRC10188)C57BL/6-Gt(ROSA)26Sor<tm1.1(CAG-3XFLAG/dCas9,-EGFP)Hfuj> (RBRC10189)C57BL/6-Gt(ROSA)26Sor<tm1.2(CAG-3XFLAG/dCas9)Hfuj> (RBRC10190)Courtesy of Hodaka Fujii, M.D., Ph.D. Structures of expression cassettes of 3xFLAG-dCas9 and a scheme of enChIP analysis using transgenic mice expressing 3xFLAG-dCas9 |
|
Molecular interaction analysis is essential to understand molecular mechanisms of genomic functions (transcription, epigenetic regulation, etc.). Engineered DNA-binding molecule-mediated chromatin immunoprecipitiation (enChIP) is a novel ChIP method with combination of CRISPR/Cas9 system [1,2]. This enables to identify associated proteins, DNA, RNA or other molecules with genomic regions of interest in a non-biased manner. The developers of enChIP analysis (Dr. Fujii and his colleagues) established useful transgenic mouse lines for enChIP analysis in primary mouse cells [3]. These strains express catalytically inactive form of Cas9 proteins of Streptococcus pyogenes which is tagged with 3xFLAG (3xFLAG-dCas9) under the control of CAG promoter in the ROSA26 locus. The experiment flow is roughly as follows: (1) design of guide RNA (gRNA) to a specific genomic locus; (2) isolation of objective cells from 3xFLAG-dCas9 expressing mice; (3) transfection/transduction of target specific gRNA for formation of gRNA and 3xFLAG-dCas9 complexes inside cells; (4) purification of the complexes by immunoprecipitiation; (5) identification of associated molecules by mass spectrometry, next-generation sequencing, and so on. The data of enChIP analysis will elucidate the unknown mechanisms of biological phenomenon and disease onset. |
| Depositor | : | Hodaka Fujii, M.D., Ph.D. Department of Biochemistry and Genome Biology, Hirosaki University Graduate School of Medicine |
|
| Strain name | : | C57BL/6-Gt(ROSA)26Sor<tm1(CAG-3XFLAG/dCas9,-EGFP)Hfuj> (3xFLAG-dCas9 or fluorescent reporter inducible expression mice by Cre/loxP and Flp/FRT system) |
|
| RBRC No. | : | RBRC10188 | |
| Strain name | : | C57BL/6-Gt(ROSA)26Sor<tm1.1(CAG-3XFLAG/dCas9,-EGFP)Hfuj> (3xFLAG-dCas9 and fluorescent reporter expression mice) |
|
| RBRC No. | : | RBRC10189 | |
| Strain name | : | C57BL/6-Gt(ROSA)26Sor<tm1.2(CAG-3XFLAG/dCas9)Hfuj> (3xFLAG-dCas9 constitutive expression mice) |
|
| RBRC No. | : | RBRC10190 | References | : | [1] | Fujita T, Fujii H. Efficient isolation of specific genomic regions and identification of associated proteins by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Biochem Biophys Res Commun.; 439(1):132-6, 2013. |
| [2] | Fujita T, Asano Y, Ohtsuka J, Takada Y, Saito K, Ohki R, Fujii H. Identification of telomere-associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Sci Rep.; 3:3171, 2013. |
||
| [3] | Fujita T, Kitaura F, Oji A, Tanigawa N, Yuno M, Ikawa M, Taniuchi I, Fujii H. Transgenic mouse lines expressing the 3xFLAG-dCas9 protein for enChIP analysis. Genes Cells.; 23(4):318-325, 2018. |
||
| Ocrober 2019 Saori Mizuno, Ph.D. Contact: Experimental Animal Division, RIKEN BioResource Research Center (animal.brc@riken.jp) All materials contained on this site may not be reproduced, distributed, displayed, published or broadcast without the prior permission of the owner of that content. |